| HELP TOPIC: "3. Uploading Entry Points" |
On This Page:
3.1. How to Upload Entry Points:
To define custom genomic entry points (e.g. chromosomes):
- Log into Genboree.
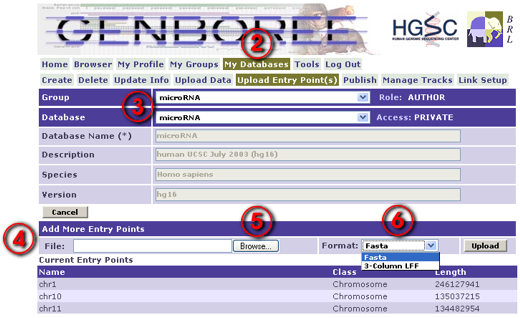
- Navigate via the menu to My Databases / Upload Entry Point(s).
- Select the appropriate Group and Database in the droplists.
- Below the database information is an interface for uploading entry points, and a list of any existing entry points.
- Select a file on your computer, select a file format, and click Upload.
- One of two file types is expected: a Fasta file or a 3-column LFF entry point file.
- Wait for the file to be transferred to the Genboree server.
- After the file is transferred, it will be queued for processing and uploading into the database. You will receive an email when the upload has been completed.
- NOTE: to upload, you must have at least the Author role in the Group.
3.2. Entry Point File Formats:
3.2.1. FASTA Sequence File
- - Use this if you have chromosomes or scaffolds in some multi-fasta files.
- - The fasta sequences will determine entry point lengths and will be available to users via the Genboree browser.
- - Genboree follows the Fasta description available at Wikipedia.
- - No attempt is made to parse application-specific unique identifiers.
- - Fasta comment lines will be stripped.
NOTE: The unique identifier is the first word (series of non-whitespace characters) following the ">" on the defline.
It determines the entry point name and is case-sensitive.
A sensible Fasta record for "chr13" might look like:
>chr13 GTCTTTGTGTCACTGACCCCTCGATATGTCCTACGATCCCATGATATGAACTCACCAGATTTTCCAATGG AAGGGATAGGAATTCCGAGAGACAGAGAGAAAGGGAGAGAGAGAGAGAGAGAGAAAAGAAAGAGAGAGAG atcaaagaaacagagagagagagagtatatatacaaaggaaacagagggatacacacaccccccactaaa tgtgatccgaggggctattacagatctcactttgttgaagtgttgcagccaattcaaaacaaactaaaca GTCATGATTATGATGACAACGATGGCGACAACACCATNNNNNNNNNNNNNNNNNNNCATCATCATCATCA . . .
or for "Scaffold_70613" in an unassembled genome:
>Scaffold_70613 tgtgatccgaggggctattacagatctcactttgttgaagtgttgcagc TTGACCAGCAGAAATAAAGCTCTGTTCACAACCTATTTTCCACACACAT GTCATGATTATGATGACAACGATGGCGACAACACCATNNNNNNNNNNNN . . .
3.2.2. 3-Column LFF Entry Point File
- - Use this if you don't have sequences for your chromosomes or scaffolds.
- - The file format is a simple tab-delimited file with 3 columns per line:
- · the entry point name
- · the keyword "Chromosome"
- · the length of the entry point
A 3-column LFF entry point file might look like:
chr1 Chromosome 246127941 chr2 Chromosome 243615958 chr3 Chromosome 199344050 . . .
or
Scaffold10 Chromosome 474987 Scaffold100 Chromosome 300122 Scaffold1000 Chromosome 165290 Scaffold100010 Chromosome 1448 Scaffold100082 Chromosome 12132 . . .

|
© 2001-2026 Baylor College of Medicine
Bioinformatics Research Laboratory (400D Jewish Wing, MS:BCM225, 1 Baylor Plaza, Houston, TX 77030) |