| HELP TOPIC: "5. The LFF Annotation Format" |
On This Page:
See Also:
Show expanded help info?
5.1. Overview:
The Genboree LFF format is adapted from the LDAS upload format described at http://www.biodas.org/, specifically from the [ Annotations ] section.
These points are generally important:
- The LFF format is tabular; each row is a single annotation record.
- The annotation record is tab-delimited into 10 required columns, with up to 5 additional optional columns.
- Regular spaces are allowed in many columns, because tabs are different than spaces.
- NOTE: Do not use '{' or '}' characters. Due to a bug in MySQL's Java library code, in certain combinations, the data will not upload even though the data otherwise appears fine. MySQL is aware of this bug.
HINT:
- Avoid LFF files with multiple sections; an annotation file should contain only annotations.
- Use comment lines—whose 1st non-whitespace character is "#"— for example, to list column headers:
#class name type subtype chrom start stop strand phase score qStart qStop attribute-comments sequence freeform-comments
5.2. Column Descriptions:
The Genboree LFF Format has:
- Ten (10) required columns:
- - class, name, type, subtype, chrom, start, stop, strand, phase, score
- Five (5) optional columns:
- - qStart, qStop, attribute-comments, sequence, freefrom-comments
A detailed description for each column follows. For a more compact view, you can hide the Genboree context images and text using the Show expanded help info? above.
LFF Annotation Columns:
Col. #1:
class
- - Required. Short text.
- - A general 'category' for the annotation's Track.
- - e.g. "Gene Predictions", "Conservation", "Repeats", "Assembly".
- - Used to categorize annotation tracks;
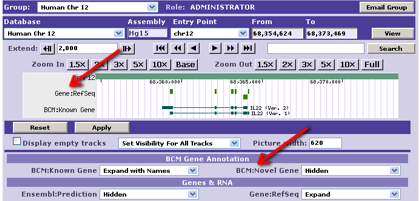
for example, below the browser picture:
Col. #2:
name
- - Required. Short text.
- - A name for the annotation/annotation group.
- - All annotations with the same name are considered grouped.
- - There are group-aware drawing styles that can suitably display such Annotation Groups.
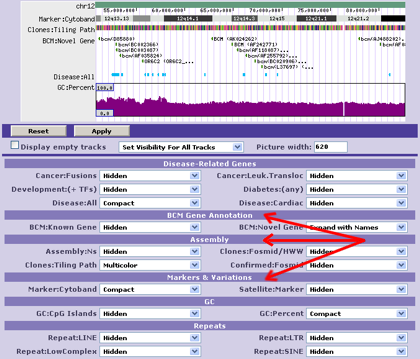
- - The exons in the 1st track all have different names and are probably not being drawn as the user would prefer.
- - The exons in the 2nd track, however, are named according to their respective gene transcripts and can be drawn sensibly.
- - Conversely, if all annotations are given the same name, they will all be in the same group. Group-aware drawing styles may not appear as you wish, and performance may suffer.
Col. #3:
type
- - Required. Very short text. E.g. name or acronymn.
- - The type of annotation; a repetition or a sensible sub-category of the class is best.
- - Actually, any text you like, as long as it doesn't contain the ':' character.
- - See "Note" under subtype below.
Col. #4:
subtype
- - Required. Very short text. E.g. name or acronymn.
- - A more specific sub-type for the annotation; something more specific than type is best.
- - Actually, any text you like, as long as it doesn't contain the ':' character.
Col. #5:
- - Required. Very short text.
- - Name of the entry point (e.g. the chromosome) the annotation is on.
- - It must be one of the entry points defined for the database.
Col. #6:
start
- - Required. Positive integer.
- - Start of annotation on the entry point.
- - Start values beyond the ends of the entry point are prohibited.
- - Note: the first base of an entry point is 1 (not 0). The start coordinate is included in the annotation.
Col. #7:
stop
- - Required. Positive integer.
- - End of annotation on the entry point.
- - Stop values beyond the ends of the entry point are prohibited.
- - Note: the first base of an entry point is 1 (not 0). The stop coordinate is included in the annotation.
Col. #8:
strand
- - Required. One of: '+' or '-'.
- - The orientation of the annotation with respect to the entry point.
- - Use '+' if you don't care about strand.
- - The strand is always available by left-clicking the annotation.
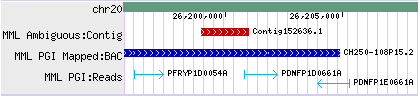
- - Some drawing styles are orientation aware:
Col. #9:
phase
- - Required. One of: 0,1,2 or '.' ('.' == n/a).
- - Whether the annotation is "in-phase" or "out-of-phase" with respect to something, such as the reading frame, or the other mate-pair read, etc.
- - Currently, one drawing style is phase-aware: Paired-End
- - Along with strand, it uses phase to visually indicate the relative orientation of mapped mate pair ends (i.e. whether the ends are in-phase or out-of-phase) when represented as a single annotation:
- → ← strand: +, phase: 0
- → → strand: +, phase: 1
- ← ← strand: -, phase: 1
- ← → strand: -, phase: 0
- - The Paired-End drawing style does this by representing + oriented ends with a green block and - oriented ends with a yellow block:
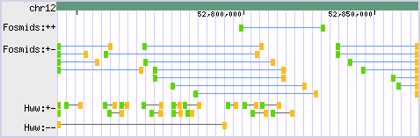
- - Other representations are possible, given user demand.
Col. #10:
score
- - Required. Real number.
- - A score for the annotation.
- - e.g. 340, 0.871, 1e-10, 0, 1.0, etc
- - We recommend "1.0" when score doesn't matter.
- - The score is always available by left-clicking the annotation.
- - Some drawing styles use the score directly.
- - The minimum/maximum is globally-derived so the y-axis scale is uniform, regardless of location/view.
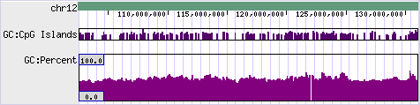
Col. #11:
qStart
- - Optional. Integer.
- - Start of hit in the query. Or '.' for n/a.
Col. #12:
qStop
- - Optional Integer.
- - Stop of hit in the query. Or '.' for n/a.
Col. #13:
attribute
comments
comments
- - Optional. A series of attribute=value; pairs.
- - The attribute names are up to you, as are the values.
- - Attribute=value; format is:
- · attribute name (any text not '=')
- · then '='
- · then value (any text not ';')
- · then ';'
- - The attribute cannot be longer than 255 characters.
- - If the value is longer than 65535, it will be truncated.
- - This column can contain multiple attribute=value; pairs.
- - Pairs found in this column are specifically modelled as 'attributes' or 'properties' of your annotation.
- - These attribute-value; pairs have additional advantages:
- self-documenting comments with a regular structure
- easy to extract data into custom Link URLs
- looks similar to other formats (i.e. GFF)
- users looking at an Annotation's Details can make use of an 'auto-wrap' feature that makes reading such comments user-friendly
- - LFF attribute-comment example:
gi=123987456; extDB_ref=10987K5; percIdent=94.68; e-val=1e-68; region=transmembrane; source=Smith Lab;
- - Comment wrapping example:
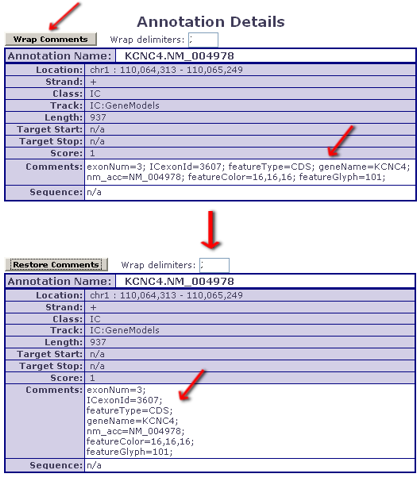
Col. #14:
sequence
- - Optional. Long text.
- - This is intended for the sequence of the query or protein mapped to this region of the genome.
- - Sometimes the query sequence and the genomic sequence are different (e.g. blating drosophila genes against sea urchin genome) and you want a place to put the query sequence.
- - Be reasonable, however; not appropriate for storing the Mouse genome.
- - Like comments, the sequence associated with an annotation will be available in the browser via left-clicking and choosing Annotation Details.
Col. #15:
freeform
comments
comments
- - Optional. Long text.
- - We strongly recommend using the attribute comments to formally record extra content. It can be used for sub-selection, custom track links, etc.
- - As a last resort, this free-form text column is provided.
- - Be reasonable, however; not appropriate for storing War and Peace.
Minimal (10 columns; a 2-exon gene):
Genes & RNA AVPR1A Gene RefSeq chr12 63256962 63258172 - . 0 . . Genes & RNA AVPR1A Gene RefSeq chr12 63260393 63263337 - . 0 . .
Standard (12 columns; contigs within a scaffold):
Assembly AAGJ01021111 Assembly Contig Scaffold_114754 1 1300 + . 1.0 1 1300 Assembly AAGJ01022222 Assembly Contig Scaffold_114754 2195 3504 - . 1.0 1 1310
With comments (13 columns; SNPs):
Cancer SNPs HUR6.188 SNPs Codon chr2 19461847 19461847 + . 0 . . allele=G/T; aaChange=A->A; nonSynon=false; refAA=A; mutAA=A; refCodon=GCG; rs_acc=rs123456; leftFlank=TGACGG; rightFlank=GCCAAC; exonPosition=2; proteinPosition=42; ampliconId=25299; Cancer SNPs HUR6.329 SNPs Codon chr12 19461988 19461988 + . 0 . . allele=C/T; aaChange=Y->Y; nonSynon=false; refAA=Y; mutAA=Y; refCodon=TAC; rs_acc=rs987654; leftFlank=ACGCGC; rightFlank=GGGCGC; exonPosition=2; proteinPosition=89; ampliconId=25299; Cancer SNPs HUR1D.382 SNPs Codon chr18 22989108 22989108 - . 0 . . allele=T/C; aaChange=N->N; nonSynon=false; refAA=N; mutAA=N; refCodon=AAT; rs_acc=rs789123; leftFlank=CTGTGT; rightFlank=GAAGAG; exonPosition=2; proteinPosition=360; ampliconId=25053; Cancer SNPs GRAF3.220 SNPs Codon chr19 36753139 36753139 - . 1 . . allele=T/G; aaChange=S->A; nonSynon=true; refAA=S; mutAA=A; refCodon=TCC; rs_acc=rs567891; leftFlank=AGCTCC; rightFlank=CCGAGT; exonPosition=7; proteinPosition=310; ampliconId=24229;
Genboree will recognize certain special attributes within the attribute comments column (13th column).
Some of these are experimental, but the generally available and stable ones are listed below.
annotationColor
- - Use this to set a color specific to this annotation.
- - The annotation-specific color will override any track color settings.
- - The color may be specified in the annotationColor attribute's value in one of 3 ways:
- RGB Hex Format:
annotationColor=#FF00AA; - RGB Dec Format:
annotationColor=255,10,128; - HTML Color Name:
annotationColor=DarkGoldenRod;
(see full list of color names)
- RGB Hex Format:

|
Genboree is built & maintained by the Bioinformatics Research Laboratory
within the Human Genome Sequencing Center at Baylor College of Medicine.
Genboree is a hosted service, but code is available
free for academic use.
|

|
|
© 2001-2025 Bioinformatics Research Laboratory
(400D Jewish Wing, MS:BCM225, 1 Baylor Plaza, Houston, TX 77030, 713-798-5433) |
|